Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Por um escritor misterioso
Descrição

Machine learning for small molecule drug discovery in academia and industry - ScienceDirect

Multisite model for P-glycoprotein drug binding. MOLCAD representation
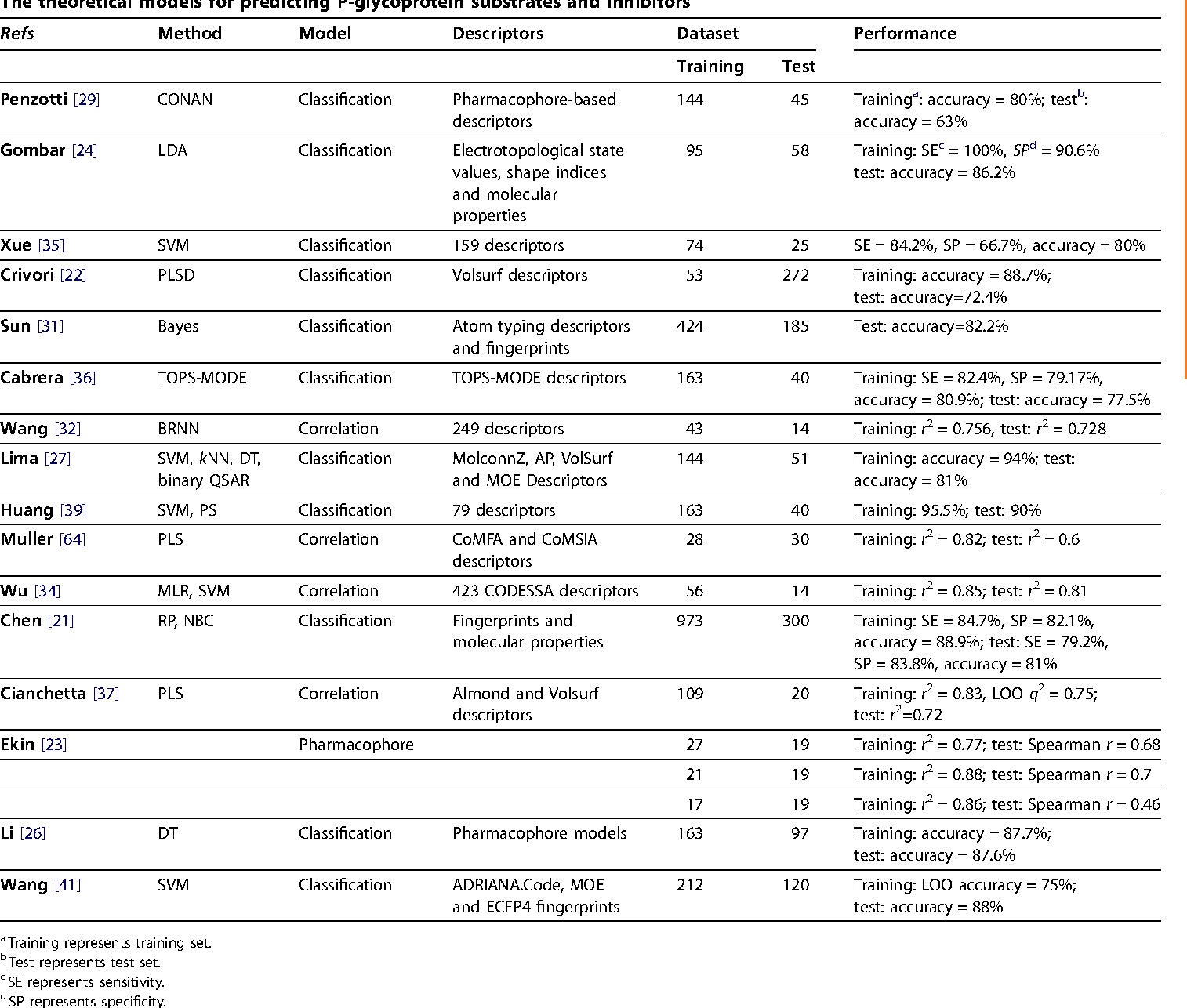
PDF] Computational models for predicting substrates or inhibitors of P- glycoprotein.
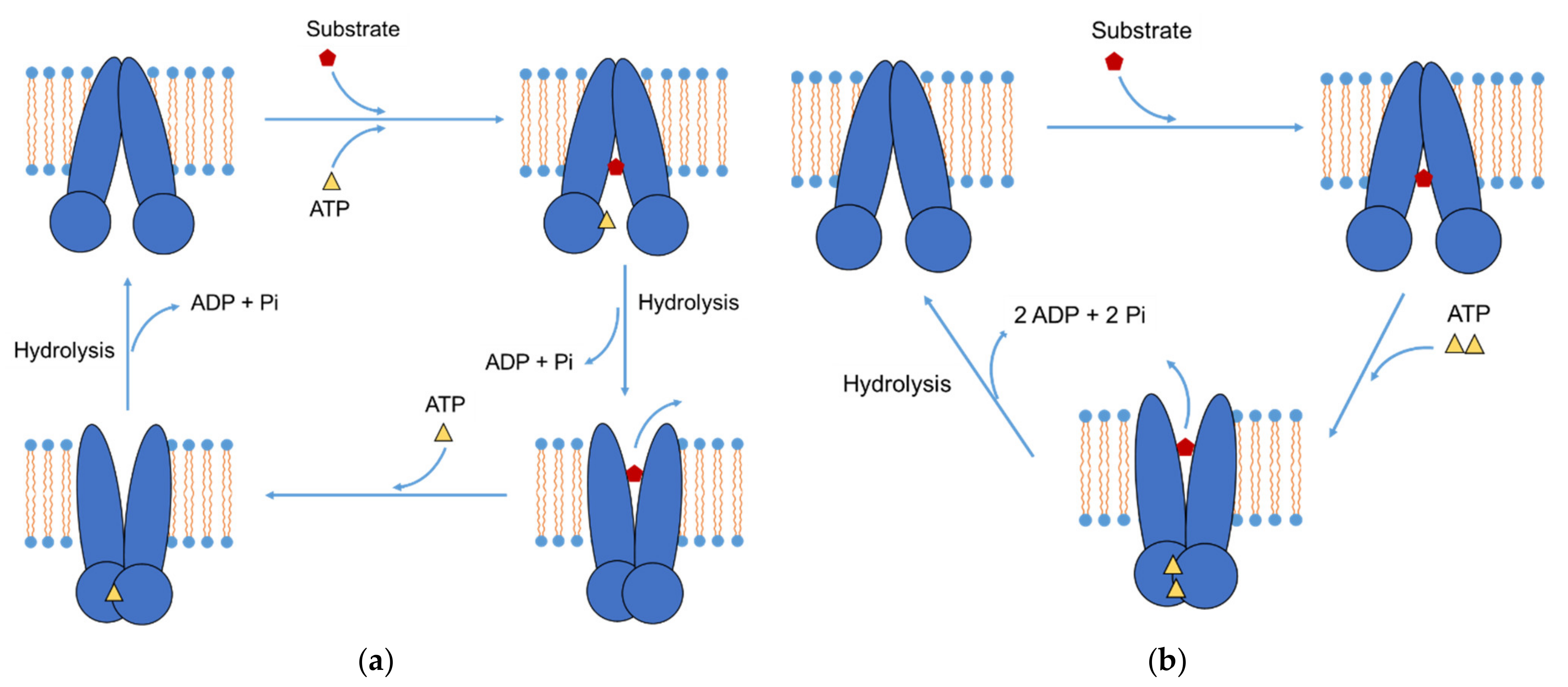
Pharmaceutics, Free Full-Text

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

Computational Biology and Chemistry in MTi: Emphasis on the Prediction of Some ADMET Properties - Miteva - 2017 - Molecular Informatics - Wiley Online Library

Full article: Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease
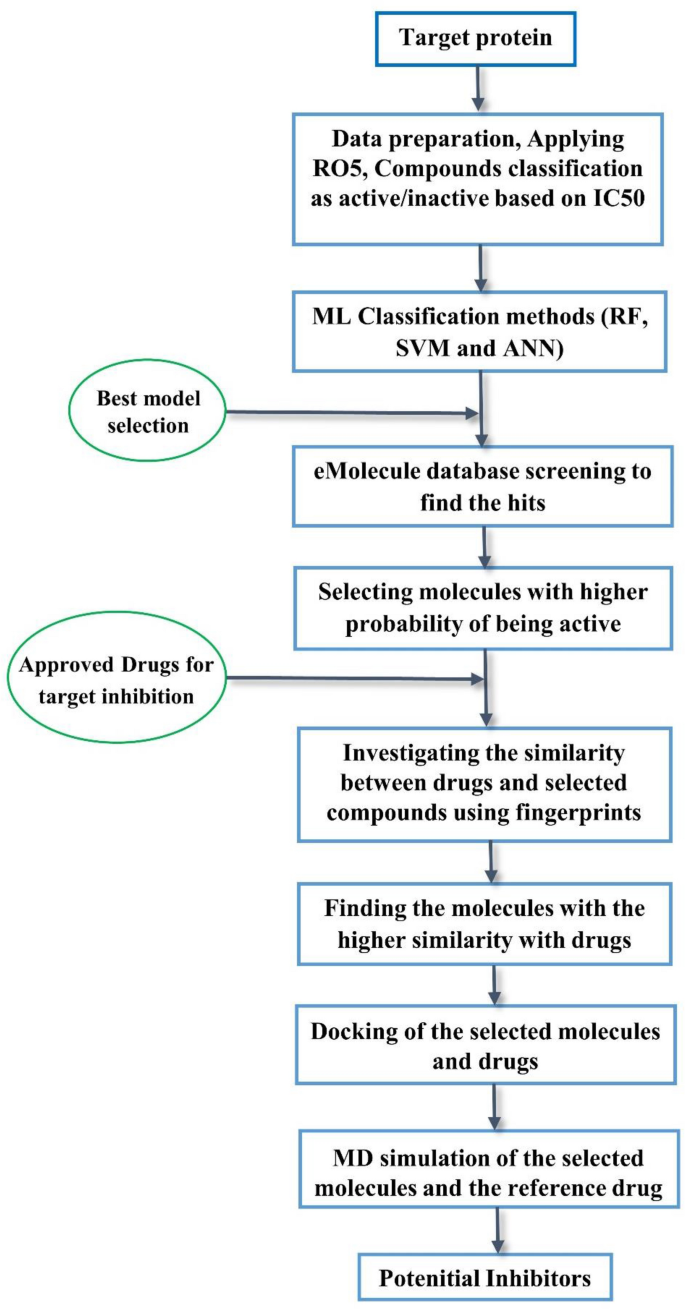
The use of machine learning modeling, virtual screening, molecular docking, and molecular dynamics simulations to identify potential VEGFR2 kinase inhibitors

Development of Simplified in Vitro P-Glycoprotein Substrate Assay and in Silico Prediction Models To Evaluate Transport Potential of P-Glycoprotein
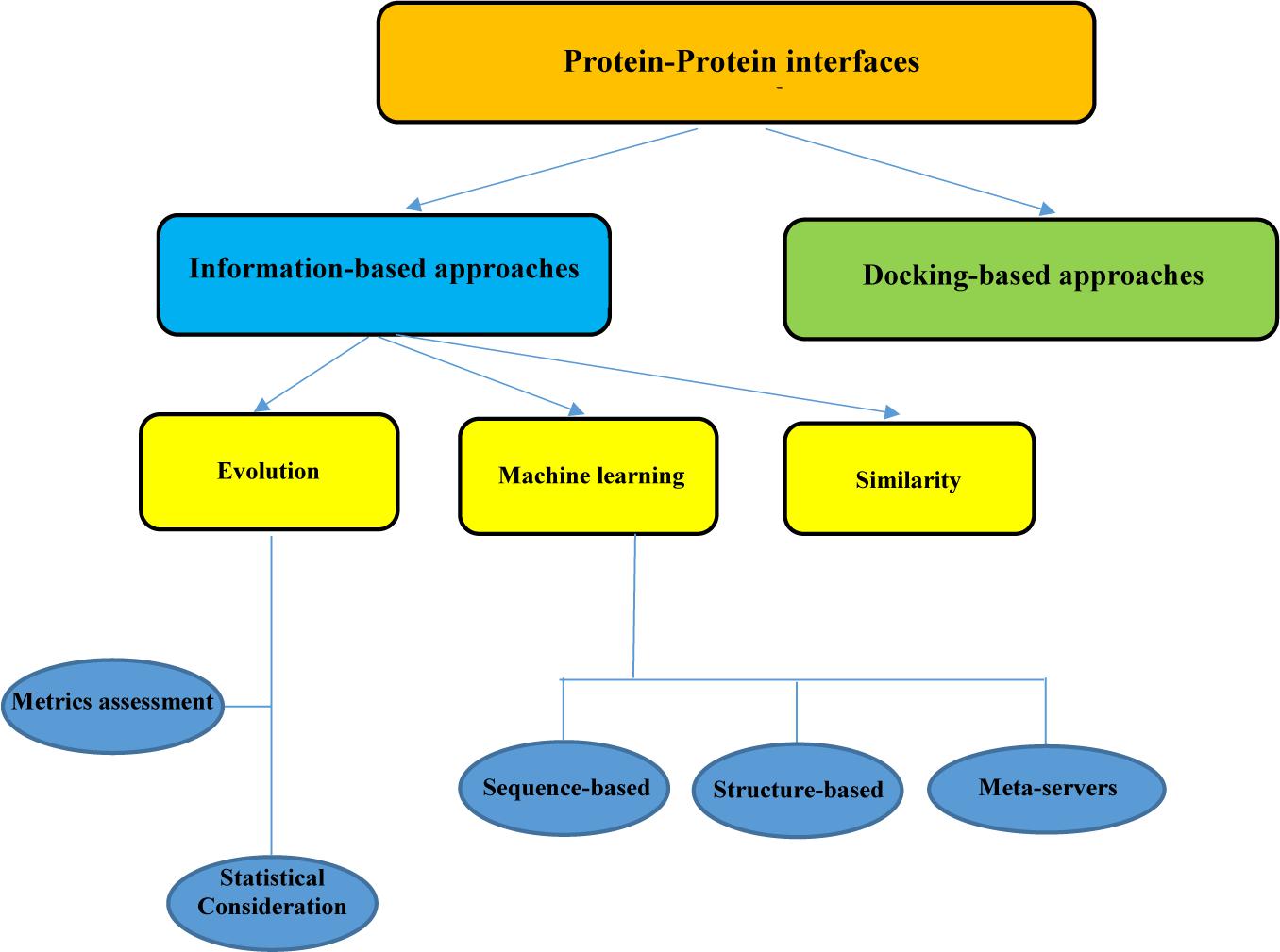
Frontiers In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein–Protein Interactions

Development of Simplified in Vitro P-Glycoprotein Substrate Assay and in Silico Prediction Models To Evaluate Transport Potential of P-Glycoprotein
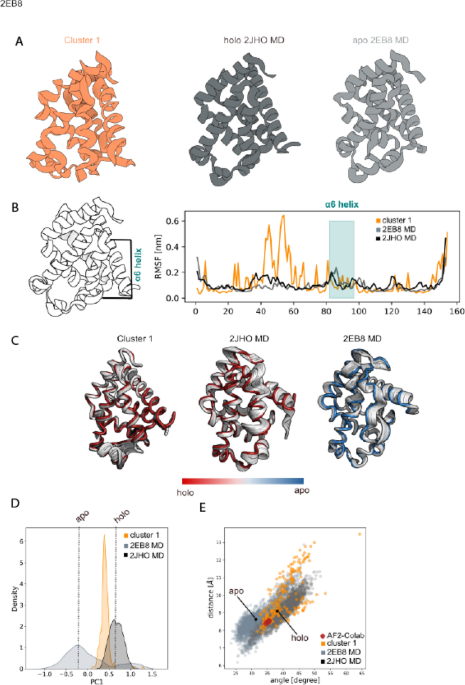
Machine learning/molecular dynamic protein structure prediction approach to investigate the protein conformational ensemble

Molecular modeling of human P-gp structure. (a) 3D Structure of human

Computational and artificial intelligence-based approaches for drug metabolism and transport prediction: Trends in Pharmacological Sciences
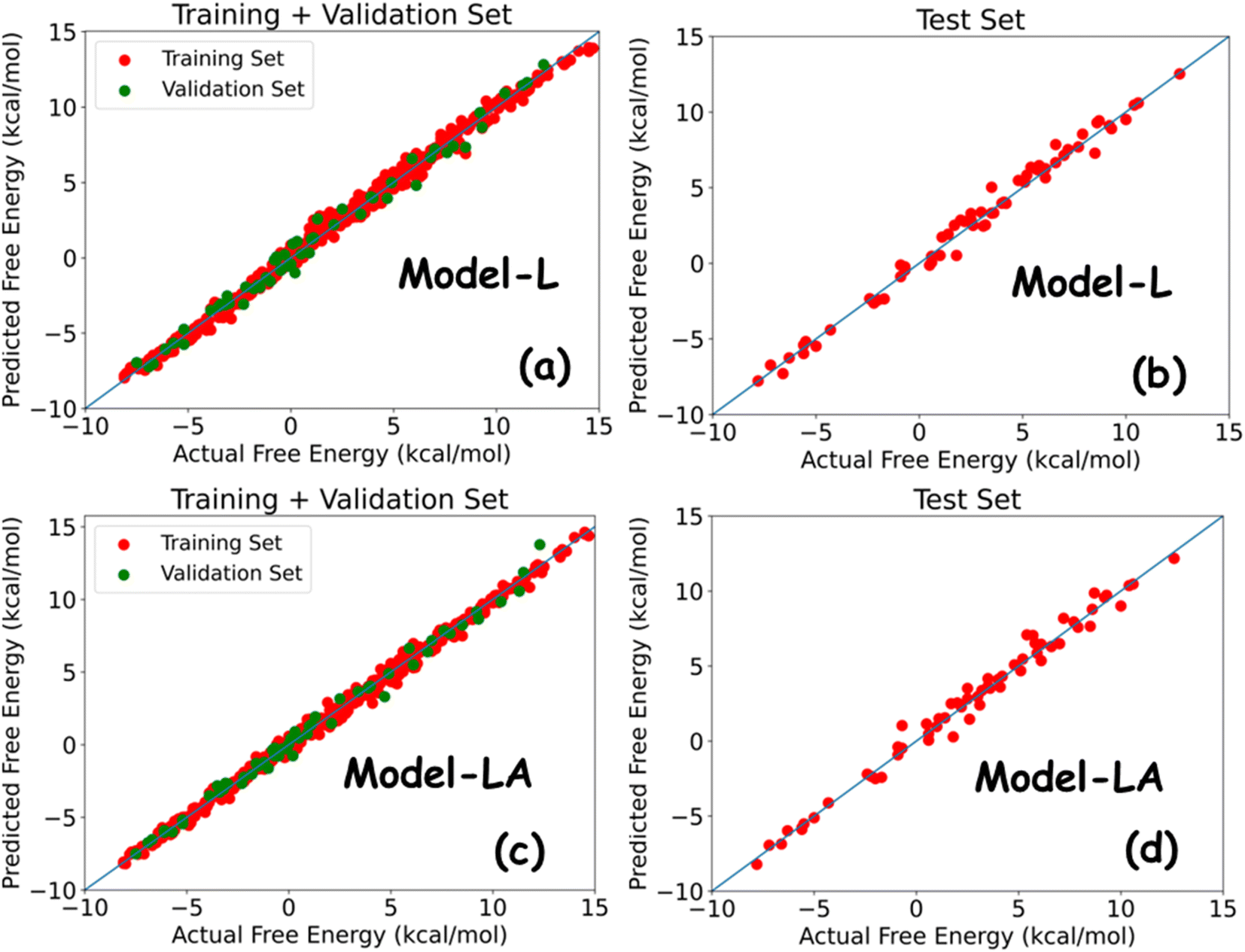
Deep learning models for the estimation of free energy of permeation of small molecules across lipid membranes - Digital Discovery (RSC Publishing) DOI:10.1039/D2DD00119E
de
por adulto (o preço varia de acordo com o tamanho do grupo)







